1. prepare data
options(stringsAsFactors = FALSE)
set.seed(999)
library("OmicCircos")
data("UCSC.hg19.chr")
data("TCGA.BC.gene.exp.2k.60")
dat <- UCSC.hg19.chr
dat$chrom <- gsub("chr", "",dat$chrom)
### initial values for simulation data
colors <- rainbow(10, alpha = 0.8)
lab.n <- 50
cnv.n <- 200
arc.n <- 30
fus.n <- 10
### make arc data
arc.d <- c()
for(i in 1:arc.n){
chr <- sample(1:19, 1)
chr.i <- which(dat$chrom == chr)
chr.arc <- dat[chr.i,]
arc.i <- sample(1:nrow(chr.arc), 2)
arc.d <- rbind(arc.d,
c(chr.arc[arc.i[1], c(1,2)],
chr.arc[arc.i[2], c(2,4)]))
}
colnames(arc.d) <- c("chr", "start", "end", "value")
### make fusion
fus.d <- c()
for(i in 1:fus.n){
chr1 <- sample(1:19, 1)
chr2 <- sample(1:19, 1)
chr1.i <- which(dat$chrom == chr1)
chr2.i <- which(dat$chrom == chr2)
chr1.f <- dat[chr1.i,]
chr2.f <- dat[chr2.i,]
fus1.i <- sample(1:nrow(chr1.f), 1)
fus2.i <- sample(1:nrow(chr2.f), 1)
n1 <- paste0("geneA", i)
n2 <- paste0("geneB", i)
fus.d <- rbind(fus.d, c(
chr1.f[fus1.i, c(1,2)], n1,
chr2.f[fus2.i, c(1,2)], n2
))
}
colnames(fus.d) <- c("chr1","po1","gene1","chr2","po2","gene2")
cnv.i <- sample(1:nrow(dat), cnv.n)
vale <- rnorm(cnv.n)
cnv.d <- data.frame(dat[cnv.i,c(1,2)], value=vale)
### gene pos
gene.pos <- TCGA.BC.gene.exp.2k.60[,1:3]
### gene expression
gene.exp <- TCGA.BC.gene.exp.2k.60
### p vale
gene.pos$p <- rnorm(250,0.01,0.001)*
sample(c(1,0.5,0.01,0.001,0.0001),250,replace=TRUE)
2. circos plot
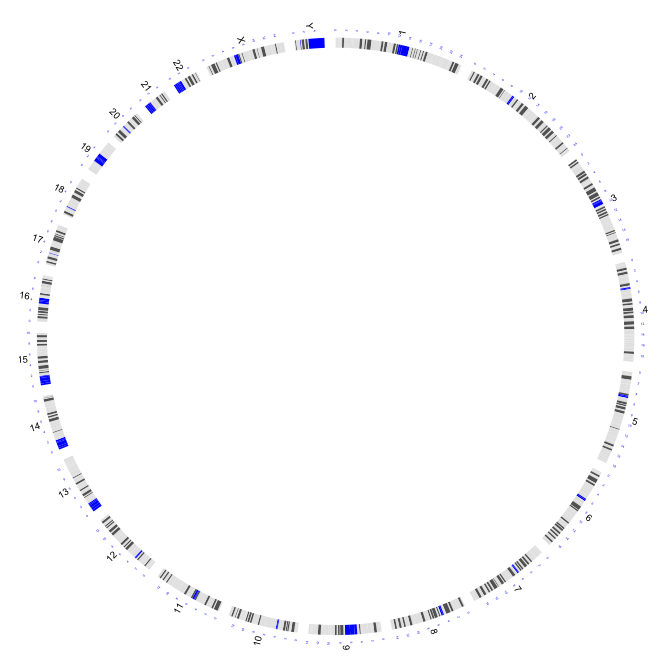
2.1 plot of chromosome
type = "chr": plots of chromosomes or segments
par(mar=c(1,1,1,1))
plot(c(1,800),c(1,800),type="n",axes=FALSE,xlab="",ylab="")
circos(R=400, cir="hg19",type="chr",W=10,scale=TRUE,print.chr.lab = TRUE)
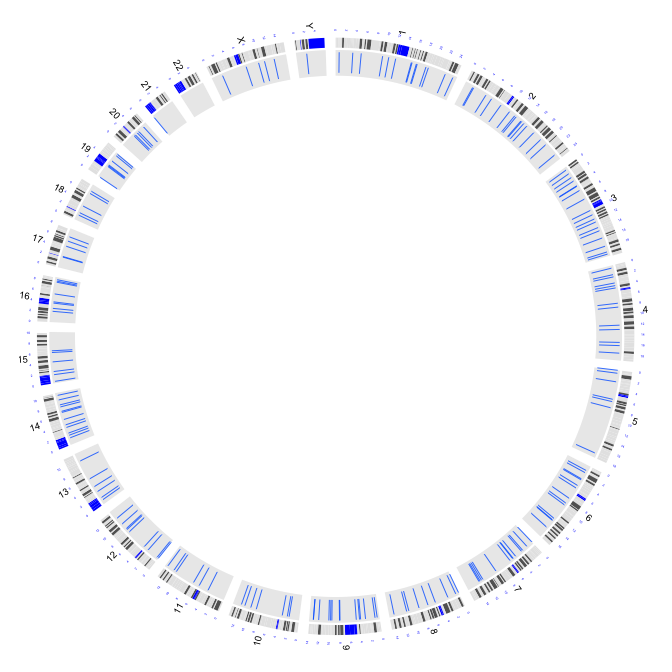
2.2 plot bar charts with the same height
type = "b3": bar charts with the same height
par(mar=c(1,1,1,1))
plot(c(1,800),c(1,800),type="n",axes=FALSE,xlab="",ylab="")
circos(R=400, cir="hg19",type="chr",W=10,scale=TRUE,print.chr.lab = TRUE)
circos(R=355, cir="hg19",type="b3",W=40,mapping=cnv.d,B=TRUE, col=colors[7])
2.3 plot dots with the fixed radius
type = "s2": dots with the fixed radius
par(mar=c(1,1,1,1))
plot(c(1,800),c(1,800),type="n",axes=FALSE,xlab="",ylab="")
circos(R=400, cir="hg19",type="chr",W=10,scale=TRUE,print.chr.lab = TRUE)
circos(R=355, cir="hg19",type="b3",W=40,mapping=cnv.d,B=TRUE, col=colors[7])
circos(R=355, cir="hg19",type="s2",W=40,mapping=cnv.d,B=FALSE, col=colors[1],cex=0.5)
2.4 plot arcs with the fixed radius
type = "arc2": arcs with the fixed radius
par(mar=c(1,1,1,1))
plot(c(1,800),c(1,800),type="n",axes=FALSE,xlab="",ylab="")
circos(R=400, cir="hg19",type="chr",W=10,scale=TRUE,print.chr.lab = TRUE)
circos(R=355, cir="hg19",type="b3",W=40,mapping=cnv.d,B=TRUE, col=colors[7])
circos(R=355, cir="hg19",type="s2",W=40,mapping=cnv.d,B=FALSE, col=colors[1],cex=0.5)
circos(R=320, cir="hg19",type="arc2",W=35,mapping=arc.d,B=TRUE, col=colors,lwd=10,cutoff=0)
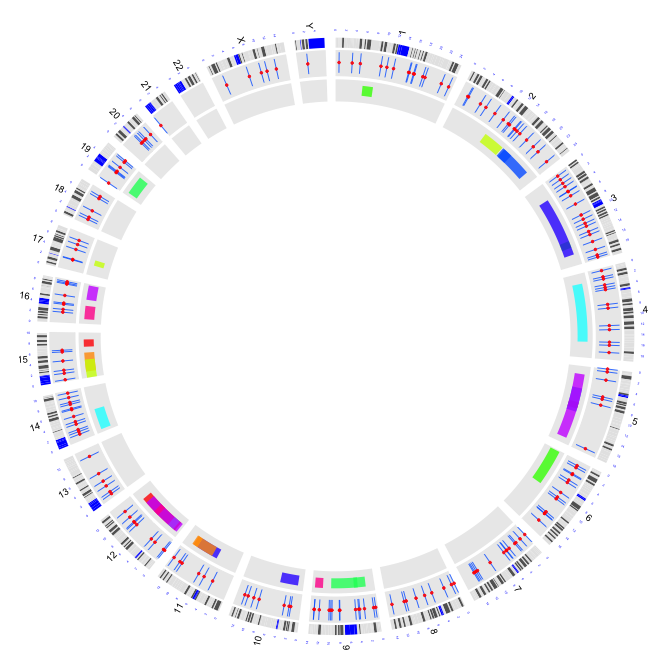
2.5 plot bar charts (opposite side of cutoff value)
type = "b2": bar charts (opposite side of cutoff value)
par(mar=c(1,1,1,1))
plot(c(1,800),c(1,800),type="n",axes=FALSE,xlab="",ylab="")
circos(R=400, cir="hg19",type="chr",W=10,scale=TRUE,print.chr.lab = TRUE)
circos(R=355, cir="hg19",type="b3",W=40,mapping=cnv.d,B=TRUE, col=colors[7])
circos(R=355, cir="hg19",type="s2",W=40,mapping=cnv.d,B=FALSE, col=colors[1],cex=0.5)
circos(R=320, cir="hg19",type="arc2",W=35,mapping=arc.d,B=TRUE, col=colors,lwd=10,cutoff=0)
circos(R=280, cir="hg19",type="b2",W=40,mapping=cnv.d,B=TRUE, col=colors[c(7,9)],lwd=2,cutoff=-0.2, col.v=3)
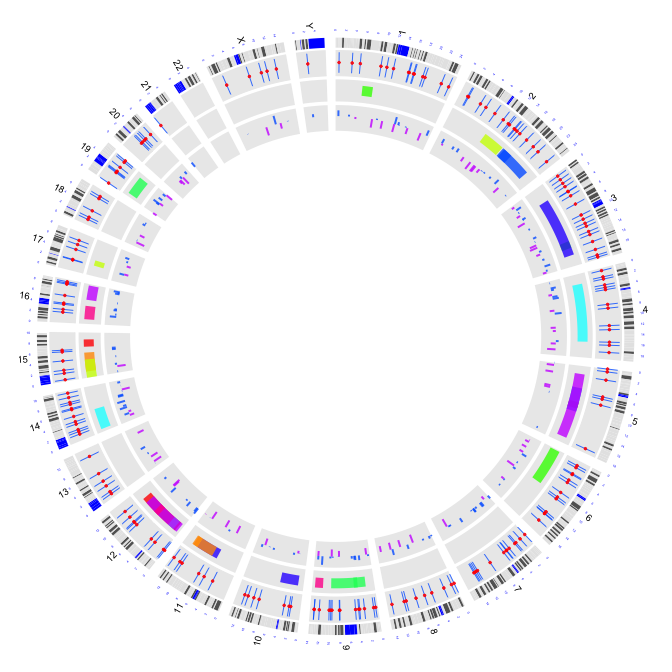
2.6 plot arcs with variable radius
type = "arc": arcs with variable radius
par(mar=c(1,1,1,1))
plot(c(1,800),c(1,800),type="n",axes=FALSE,xlab="",ylab="")
circos(R=400, cir="hg19",type="chr",W=10,scale=TRUE,print.chr.lab = TRUE)
circos(R=355, cir="hg19",type="b3",W=40,mapping=cnv.d,B=TRUE, col=colors[7])
circos(R=355, cir="hg19",type="s2",W=40,mapping=cnv.d,B=FALSE, col=colors[1],cex=0.5)
circos(R=320, cir="hg19",type="arc2",W=35,mapping=arc.d,B=TRUE, col=colors,lwd=10,cutoff=0)
circos(R=280, cir="hg19",type="b2",W=40,mapping=cnv.d,B=TRUE, col=colors[c(7,9)],lwd=2,cutoff=-0.2, col.v=3)
circos(R=240, cir="hg19",type="arc",W=40,mapping=arc.d,B=TRUE, col=colors[c(1,7)],lwd=4,scale = TRUE,col.v=4)
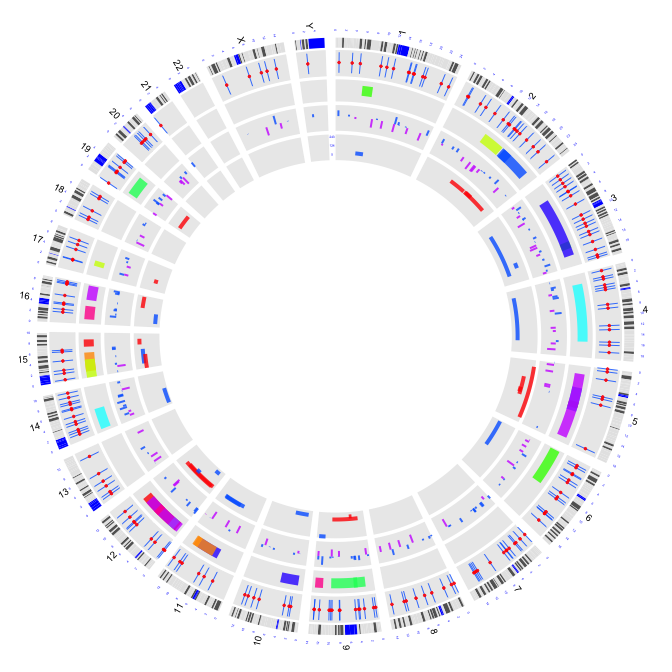
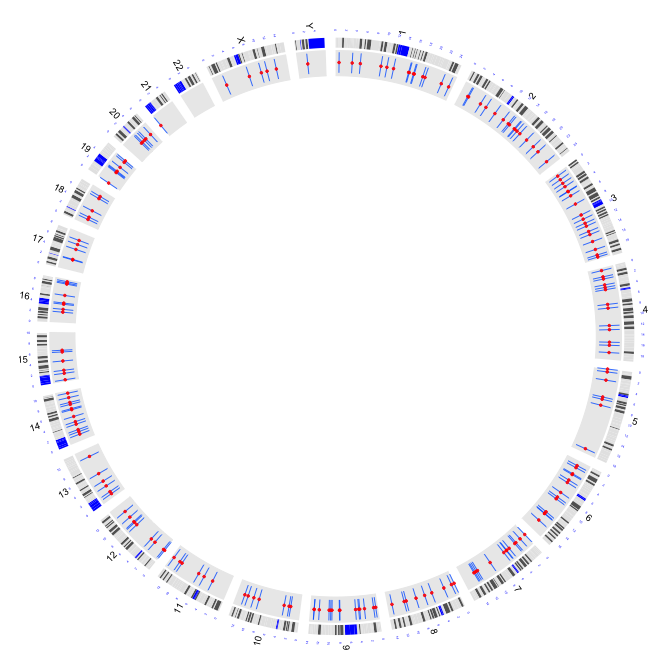
2.7 box plots
par(mar=c(1,1,1,1))
plot(c(1,800),c(1,800),type="n",axes=FALSE,xlab="",ylab="")
circos(R=400, cir="hg19",type="chr",W=10,scale=TRUE,print.chr.lab = TRUE)
circos(R=355, cir="hg19",type="b3",W=40,mapping=cnv.d,B=TRUE, col=colors[7])
circos(R=355, cir="hg19",type="s2",W=40,mapping=cnv.d,B=FALSE, col=colors[1],cex=0.5)
circos(R=320, cir="hg19",type="arc2",W=35,mapping=arc.d,B=TRUE, col=colors,lwd=10,cutoff=0)
circos(R=280, cir="hg19",type="b2",W=40,mapping=cnv.d,B=TRUE, col=colors[c(7,9)],lwd=2,cutoff=-0.2, col.v=3)
circos(R=240, cir="hg19",type="arc",W=40,mapping=arc.d,B=TRUE, col=colors[c(1,7)],lwd=4,scale = TRUE,col.v=4)
circos(R=200, cir="hg19",type="box",W=40,mapping=cnv.d,B=TRUE, col=colors[1],lwd=0.1,scale = TRUE,col.v = 3)
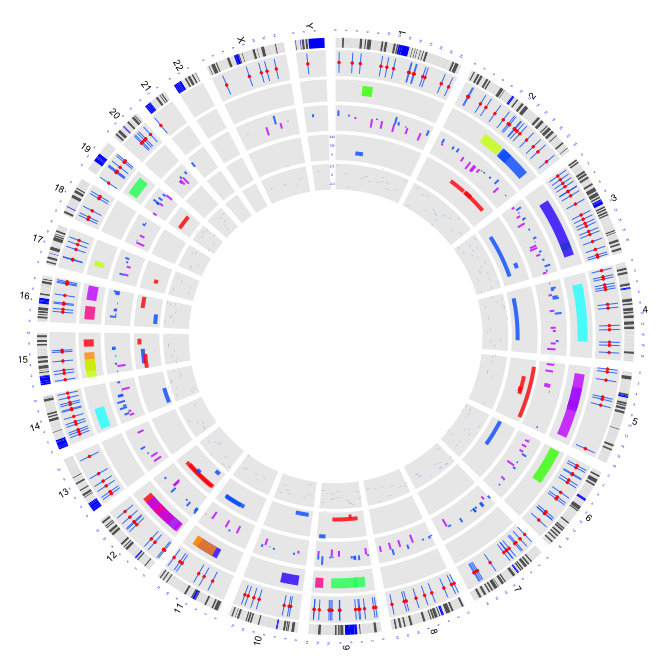
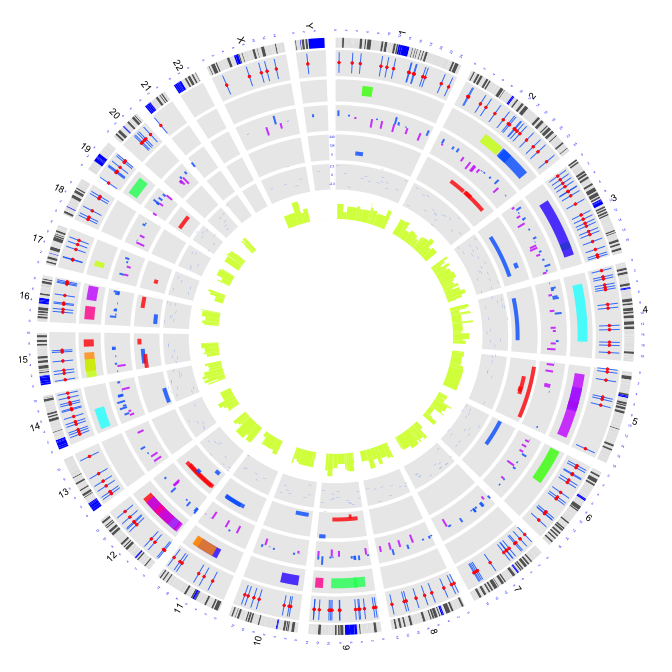
2.8 histograms
par(mar=c(1,1,1,1))
plot(c(1,800),c(1,800),type="n",axes=FALSE,xlab="",ylab="")
circos(R=400, cir="hg19",type="chr",W=10,scale=TRUE,print.chr.lab = TRUE)
circos(R=355, cir="hg19",type="b3",W=40,mapping=cnv.d,B=TRUE, col=colors[7])
circos(R=355, cir="hg19",type="s2",W=40,mapping=cnv.d,B=FALSE, col=colors[1],cex=0.5)
circos(R=320, cir="hg19",type="arc2",W=35,mapping=arc.d,B=TRUE, col=colors,lwd=10,cutoff=0)
circos(R=280, cir="hg19",type="b2",W=40,mapping=cnv.d,B=TRUE, col=colors[c(7,9)],lwd=2,cutoff=-0.2, col.v=3)
circos(R=240, cir="hg19",type="arc",W=40,mapping=arc.d,B=TRUE, col=colors[c(1,7)],lwd=4,scale = TRUE,col.v=4)
circos(R=200, cir="hg19",type="box",W=40,mapping=cnv.d,B=TRUE, col=colors[1],lwd=0.1,scale = TRUE,col.v=3)
circos(R=160, cir="hg19",type="h",W=40,mapping=cnv.d,B=FALSE, col=colors[3],lwd=0.1,col.v=3)
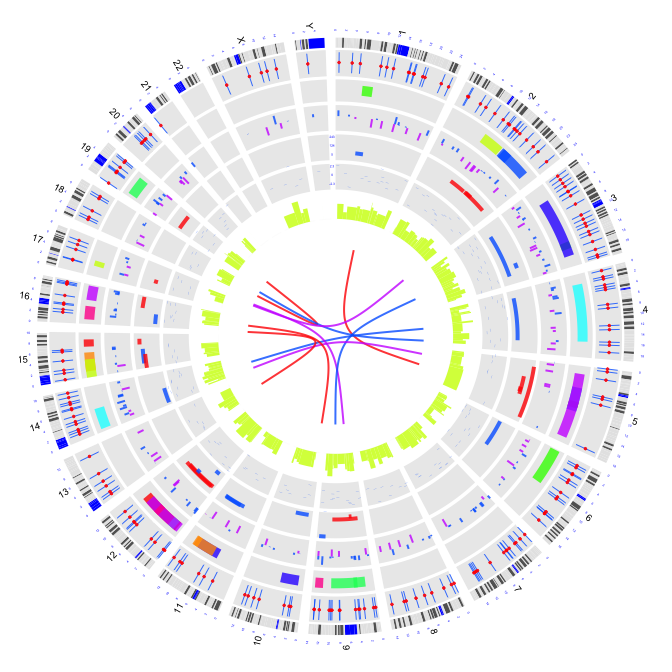
2.9 plot link lines based on Bezier curve
type = "link": link lines based on Bezier curve
par(mar=c(1,1,1,1))
plot(c(1,800),c(1,800),type="n",axes=FALSE,xlab="",ylab="")
circos(R=400, cir="hg19",type="chr",W=10,scale=TRUE,print.chr.lab = TRUE)
circos(R=355, cir="hg19",type="b3",W=40,mapping=cnv.d,B=TRUE, col=colors[7])
circos(R=355, cir="hg19",type="s2",W=40,mapping=cnv.d,B=FALSE, col=colors[1],cex=0.5)
circos(R=320, cir="hg19",type="arc2",W=35,mapping=arc.d,B=TRUE, col=colors,lwd=10,cutoff=0)
circos(R=280, cir="hg19",type="b2",W=40,mapping=cnv.d,B=TRUE, col=colors[c(7,9)],lwd=2,cutoff=-0.2, col.v=3)
circos(R=240, cir="hg19",type="arc",W=40,mapping=arc.d,B=TRUE, col=colors[c(1,7)],lwd=4,scale = TRUE,col.v=4)
circos(R=200, cir="hg19",type="box",W=40,mapping=cnv.d,B=TRUE, col=colors[1],lwd=0.1,scale = TRUE,col.v=3)
circos(R=160, cir="hg19",type="h",W=40,mapping=cnv.d,B=FALSE, col=colors[3],lwd=0.1,col.v=3)
circos(R=120,cir="hg19",type="link",W=10,mapping=fus.d,col=colors[c(1,7,9)],lwd=2)
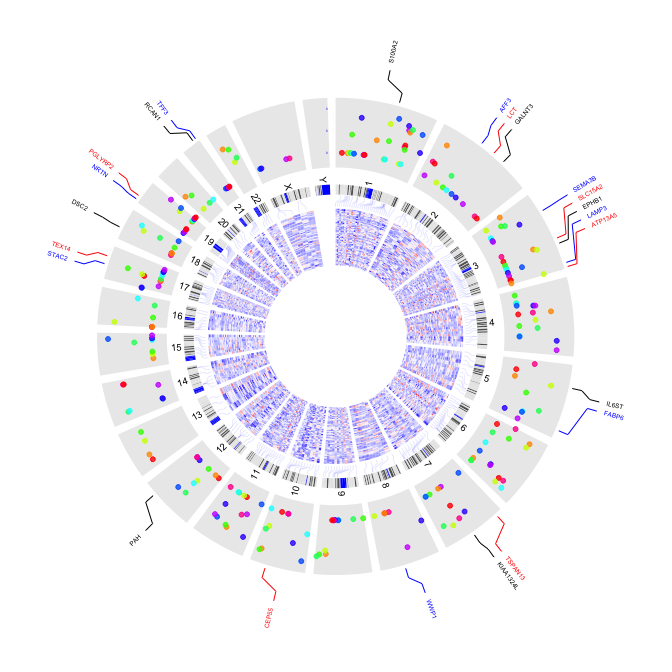
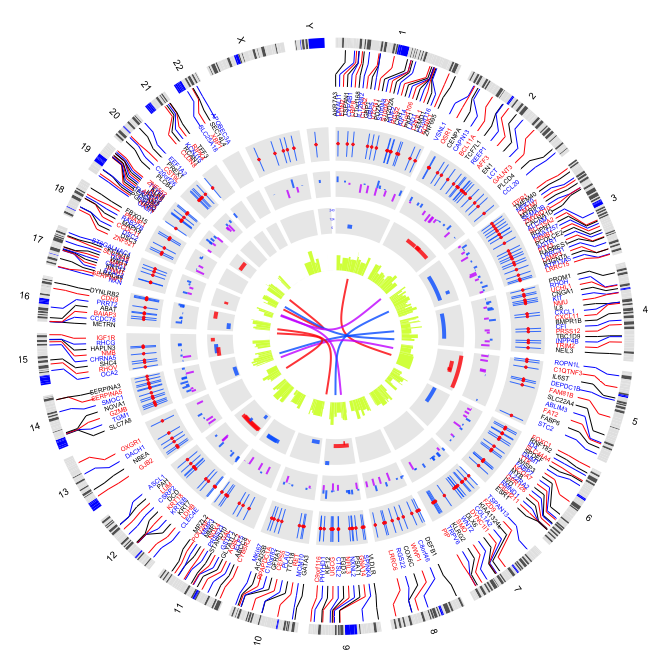
3 plot label
3.1 outside label
par(mar=c(1,1,1,1))
plot(c(1,800),c(1,800),type="n",axes=FALSE,xlab="",ylab="")
circos(R=300, cir="hg19",type="chr",W=10,scale=FALSE,print.chr.lab = FALSE)
circos(R=310, cir="hg19",type="label",W=40,mapping=gene.pos, col=c("black","blue","red"),cex=0.4,side="out")
circos(R=250, cir="hg19",type="b3",W=40,mapping=cnv.d,B=TRUE, col=colors[7])
circos(R=250, cir="hg19",type="s2",W=40,mapping=cnv.d,B=FALSE, col=colors[1],cex=0.5)
circos(R=220, cir="hg19",type="arc2",W=30,mapping=arc.d,B=TRUE, col=colors,lwd=10,cutoff=0)
circos(R=190, cir="hg19",type="b2",W=30,mapping=cnv.d,B=TRUE, col=colors[c(7,9)],lwd=2,cutoff=-0.2, col.v=3)
circos(R=160, cir="hg19",type="arc",W=30,mapping=arc.d,B=TRUE, col=colors[c(1,7)],lwd=4,scale = TRUE,col.v=4)
circos(R=130, cir="hg19",type="box",W=30,mapping=cnv.d,B=TRUE, col=colors[1],lwd=0.1,scale = TRUE,col.v=3)
circos(R=100, cir="hg19",type="h",W=30,mapping=cnv.d,B=FALSE, col=colors[3],lwd=0.1,col.v=3)
circos(R=90,cir="hg19",type="link",mapping=fus.d,col=colors[c(1,7,9)],lwd=2)
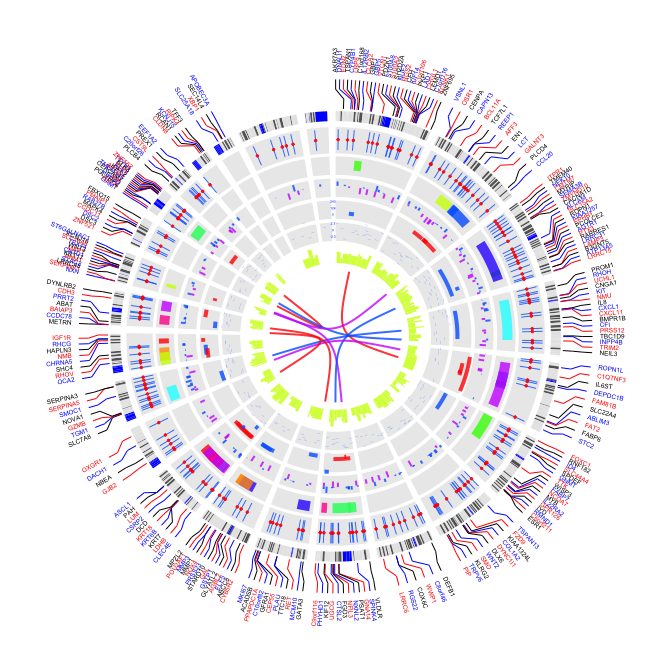
3.2 inside label
par(mar=c(1,1,1,1))
plot(c(1,800),c(1,800),type="n",axes=FALSE,xlab="",ylab="")
circos(R=400, cir="hg19",type="chr",W=10,scale=FALSE,print.chr.lab = T)
circos(R=390, cir="hg19",type="label",W=50,mapping=gene.pos, col=c("black","blue","red"),cex=0.4,side="in")
circos(R=240, cir="hg19",type="b3",W=50,mapping=cnv.d,B=TRUE, col=colors[7])
circos(R=240, cir="hg19",type="s2",W=50,mapping=cnv.d,B=FALSE, col=colors[1],cex=0.5)
circos(R=190, cir="hg19",type="b2",W=40,mapping=cnv.d,B=TRUE, col=colors[c(7,9)],lwd=2,cutoff=-0.2, col.v=3)
circos(R=140, cir="hg19",type="arc",W=40,mapping=arc.d,B=TRUE, col=colors[c(1,7)],lwd=4,scale = TRUE,col.v=4)
circos(R=90, cir="hg19",type="h",W=40,mapping=cnv.d,B=FALSE, col=colors[3],lwd=0.1,col.v=3)
circos(R=80,cir="hg19",type="link",mapping=fus.d,col=colors[c(1,7,9)],lwd=2)
4 heatmap
par(mar=c(1,1,1,1))
plot(c(1,800),c(1,800),type="n",axes=FALSE,xlab="",ylab="")
circos(R=400, cir="hg19",type="chr",W=10,scale=FALSE,print.chr.lab = T)
circos(R=300, cir="hg19",type="heatmap2",W=100,mapping=gene.exp,
col.v=4, cluster=FALSE,col.bar = FALSE,lwd=0.1,col="blue")
circos(R=200, cir="hg19",type="s",W=100,mapping = gene.pos,
col.v=4,col=colors,scale = TRUE,B=TRUE)
sig.gene <- gene.pos[gene.pos$p<0.000001,]
circos(R=190, cir="hg19",type="label",W=40,mapping=sig.gene, col=c("black","blue","red"),cex=0.4,side="in")
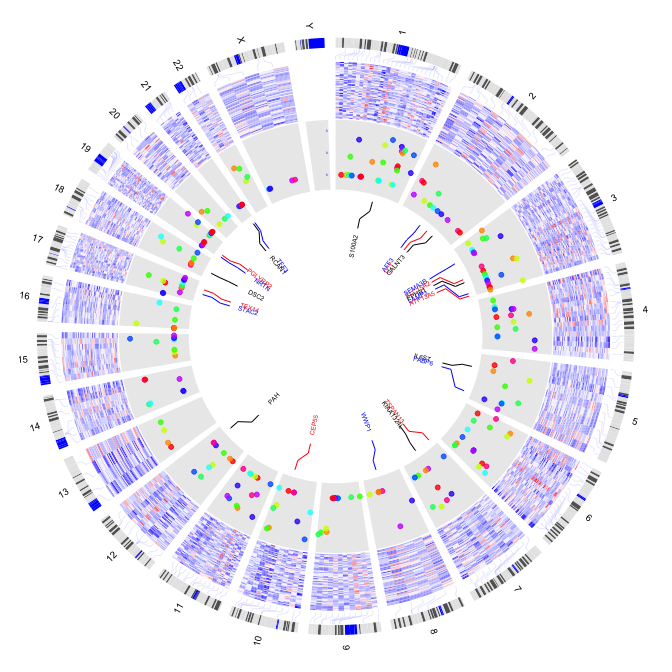
par(mar=c(1,1,1,1))
plot(c(1,800),c(1,800),type="n",axes=FALSE,xlab="",ylab="")
circos(R=200, cir="hg19",type="chr",W=10,scale=FALSE,print.chr.lab = T)
circos(R=100, cir="hg19",type="heatmap2",W=100,mapping=gene.exp,
col.v=4, cluster=FALSE,col.bar = FALSE,lwd=0.1,col="blue")
circos(R=230, cir="hg19",type="s",W=100,mapping = gene.pos,
col.v=4,col=colors,scale = TRUE,B=TRUE)
sig.gene <- gene.pos[gene.pos$p<0.000001,]
circos(R=330, cir="hg19",type="label",W=40,mapping=sig.gene, col=c("black","blue","red"),cex=0.4,side="out")