If alleles A and B are the coupled alleles at two different loci and if \(p_{B} = p_{A} + v\), with v = 0, and if \(r^2\) exceeds some threshold t, then
\[ v_{min} = \frac{p_{A}(1 - p_{A})(1 - t)}{(1 - p_{A}(1 - t))} \] \[ v_{max} = \frac{p_{A}(1 - p_{A})(1 - t)}{(p_{A}(1 - t) + t)} \]
library(plyr)
pA <- seq(0.001,0.999,0.001)
pa <- 1-pA
t2 <- rep(0.2,999)
t5 <- rep(0.5,999)
t8 <- rep(0.8,999)
df <- data.frame(pA=pA,pa=pa,t2=t2,t5=t5,t8=t8)
df2 <- transform(df,
vmax2=pa*pA*(1-t2)/(pA*(1-t2)+t2),
vmax5=pa*pA*(1-t5)/(pA*(1-t5)+t5),
vmax8=pa*pA*(1-t8)/(pA*(1-t8)+t8),
vmin2=pa*pA*(1-t2)/(1-pA*(1-t2)),
vmin5=pa*pA*(1-t5)/(1-pA*(1-t5)),
vmin8=pa*pA*(1-t8)/(1-pA*(1-t8))
)
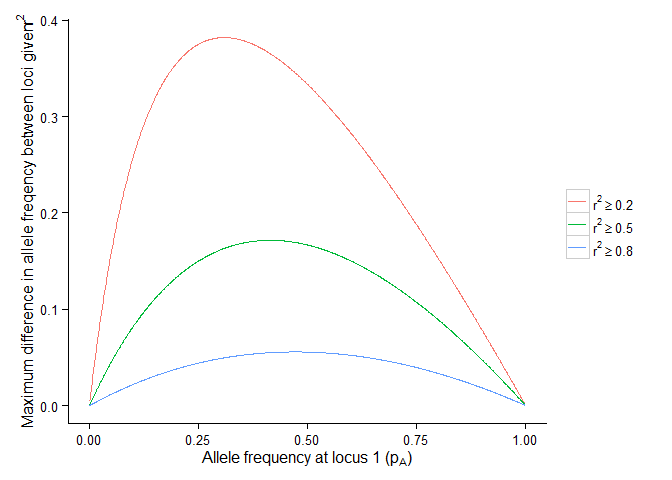
Maximum difference \(v_{max}\) between allele frequency between loci given \(r^2\)
library(ggplot2)
library(reshape2)
df <- df2[,c("pA","vmax2","vmax5","vmax8")]
df <-melt(df, id="pA")
ggplot(df,aes(x=pA,y=value,colour=variable))+
geom_line()+
theme_bw() +
theme(panel.border = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
axis.line = element_line(colour = "black"),
legend.title=element_blank())+
labs(x = expression("Allele frequency at locus 1 (p"[A]*")"),
y = expression(paste("Maximum difference in allele freqency between loci given\t", r^2)))+
scale_colour_discrete(breaks=c("vmax2", "vmax5", "vmax8"),
labels=c(expression(r^2 >= 0.2),
expression(r^2 >= 0.5),
expression(r^2 >= 0.8)))
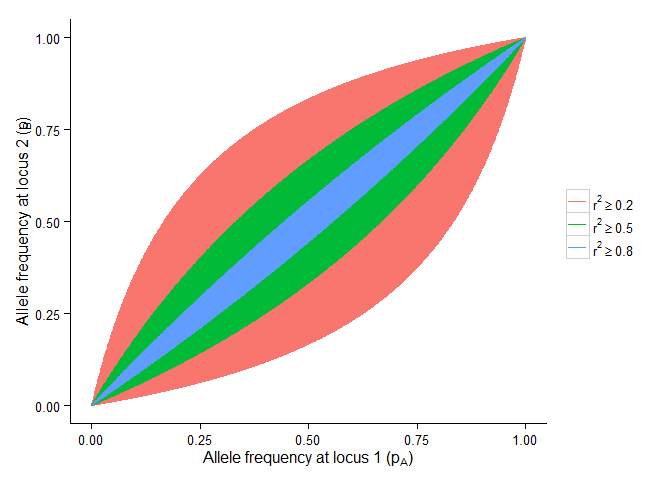
Possible range of allele frequencies at two loci given the LD between the two loci
df3 <- transform(df2,pB2=pA+vmax2,pB5=pA+vmax5,pB8=pA+vmax8)
df4 <- transform(df2,pB2=pA-vmin2,pB5=pA-vmin5,pB8=pA-vmin8)
df <- rbind(df3,df4)
df <- df[,c("pA","pB2","pB5","pB8")]
df <-melt(df, id="pA")
ggplot(df,aes(x=pA,y=value,colour=variable))+
geom_line()+
theme_bw() +
theme(panel.border = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
axis.line = element_line(colour = "black"),
legend.title=element_blank())+
labs(x = expression("Allele frequency at locus 1 (p"[A]*")"),
y = expression("Allele frequency at locus 2 (p"[B]*")"))+
scale_colour_discrete(breaks=c("pB2", "pB5", "pB8"),
labels=c(expression(r^2 >= 0.2),
expression(r^2 >= 0.5),
expression(r^2 >= 0.8)))
reference
- Allele frequencies and the r2 measure of linkage disequilibrium: impact on design and interpretation of association studies. Twin Res Hum Genet. 2005 Apr;8(2):87-94.