library(kinship2)
data(sample.ped)
sample.ped2 = subset(sample.ped, ped == 2)
sample.ped2
ped id father mother sex affected avail
42 2 201 0 0 1 1 1
43 2 202 0 0 2 NA 0
44 2 203 0 0 1 1 1
45 2 204 201 202 2 0 1
46 2 205 201 202 1 NA 0
47 2 206 201 202 2 1 1
48 2 207 201 202 2 1 1
49 2 208 201 202 2 0 0
50 2 209 0 0 1 0 0
51 2 210 203 204 1 0 0
52 2 211 203 204 1 0 1
53 2 212 209 208 2 0 1
54 2 213 209 208 1 0 0
55 2 214 209 208 1 1 1
Simple plot
pedAll <- pedigree(id = sample.ped2$id, dadid = sample.ped2$father, momid = sample.ped2$mother,
sex = sample.ped2$sex, famid = sample.ped2$ped)
print(pedAll)
Pedigree list with 14 total subjects in 1 families
print(pedAll["2"])
Pedigree object with 14 subjects, family id= 2
Bit size= 16
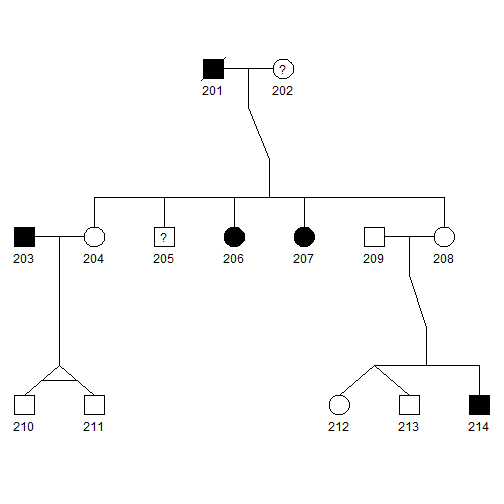
plot(pedAll["2"])
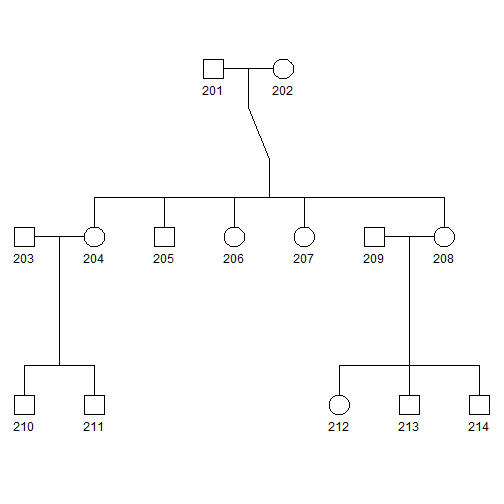
Since there is only one family, or
pedAll <- pedigree(id = sample.ped2$id, dadid = sample.ped2$father, momid = sample.ped2$mother,
sex = sample.ped2$sex, )
print(pedAll)
Pedigree object with 14 subjects
Bit size= 16
plot(pedAll)
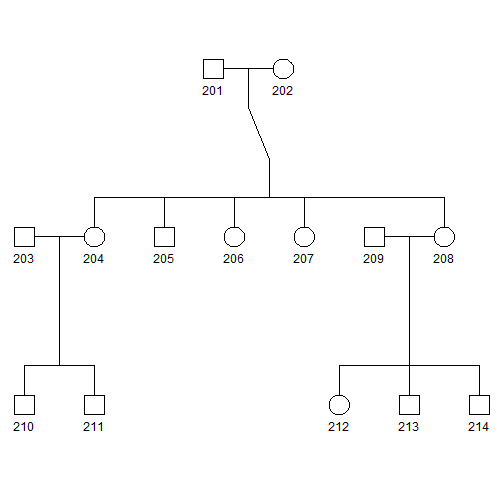
- Squares and circles indicate males and females, respectively.
Add affection status
pedAll <- pedigree(id = sample.ped2$id, dadid = sample.ped2$father, momid = sample.ped2$mother,
sex = sample.ped2$sex, affected = sample.ped2$affected)
print(pedAll)
Pedigree object with 14 subjects
Bit size= 16
plot(pedAll)
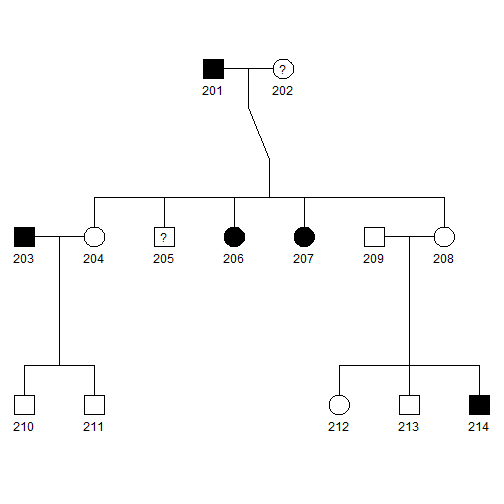
- Blackened squares and circles indicate the affected males and females, respectively.
- Squares and circles with a central question mark represent affection status is unknown.
Add status (0=”censored”, 1=”dead”)
Suppose the first individual (ID=201) is dead.
sample.ped2$status = c(1, rep(0, 13))
pedAll <- pedigree(id = sample.ped2$id, dadid = sample.ped2$father, momid = sample.ped2$mother,
sex = sample.ped2$sex, affected = sample.ped2$affected, status = sample.ped2$status)
print(pedAll)
Pedigree object with 14 subjects
Bit size= 16
plot(pedAll)
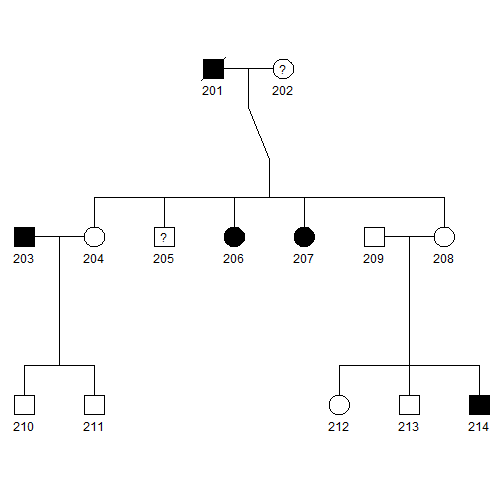
- Squares and circles with a slash represent deceased.
Add relationship (1=Monozygotic twin, 2=Dizygotic twin, 3=Twin of unknown zygosity, 4=Spouse)
Suppose 210 and 211 are monozygotic twin, 212 and 213 are dizygotic twin, create a matrix with 3 columns (id1, id2, code) specifying special relationship between pairs of individuals.
id1 = c(210, 212)
id2 = c(211, 213)
r = c(1, 2)
rm = cbind(id1, id2, r)
pedAll <- pedigree(id = sample.ped2$id, dadid = sample.ped2$father, momid = sample.ped2$mother,
sex = sample.ped2$sex, affected = sample.ped2$affected, status = sample.ped2$status,
relation = rm)
print(pedAll)
Pedigree object with 14 subjects
Bit size= 16
plot(pedAll)