The gplots package provides Venn diagrams for up to five sets. Its input is a table that is produced by another function. The function venn() calls one after the other and is the only one to be seen by the user. The venn() function accepts either a list of sets as an argument, or it takes a binary matrix, one column per set, indicating for every element, one per row, the membership with every set.
library(gplots)
venn(list(A = 1:5, B = 4:6, C = c(4, 8:10)))
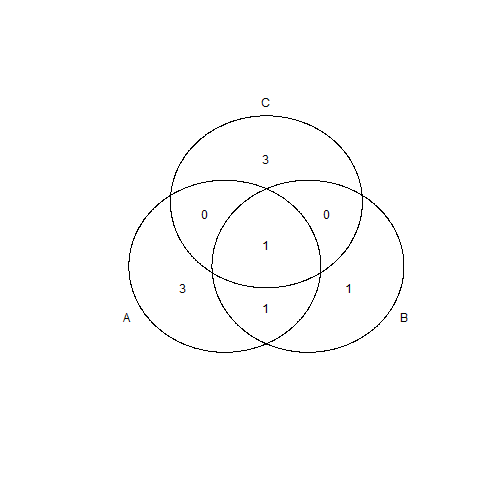
The names of columns or the list elements are the set names. To squeeze extra circles in, those circles need to become ellipses. This works for four sets and maybe even more impressively also for five.
v.table <- venn(list(A = 1:5, B = 4:6, C = c(4, 8:10), D = c(4:12)))
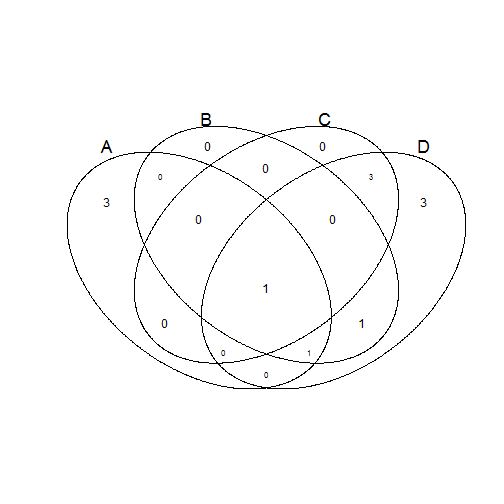
print(v.table)
num A B C D
0000 0 0 0 0 0
0001 3 0 0 0 1
0010 0 0 0 1 0
0011 3 0 0 1 1
0100 0 0 1 0 0
0101 1 0 1 0 1
0110 0 0 1 1 0
0111 0 0 1 1 1
1000 3 1 0 0 0
1001 0 1 0 0 1
1010 0 1 0 1 0
1011 0 1 0 1 1
1100 0 1 1 0 0
1101 1 1 1 0 1
1110 0 1 1 1 0
1111 1 1 1 1 1
attr(,"class")
[1] "venn"
## construct some fake gene names..
oneName <- function() paste(sample(LETTERS, 5, replace = TRUE), collapse = "")
geneNames <- replicate(1000, oneName())
##
GroupA <- sample(geneNames, 400, replace = FALSE)
GroupB <- sample(geneNames, 750, replace = FALSE)
GroupC <- sample(geneNames, 250, replace = FALSE)
GroupD <- sample(geneNames, 300, replace = FALSE)
input <- list(GA = GroupA, GB = GroupB, GC = GroupC, GD = GroupD)
venn(input)
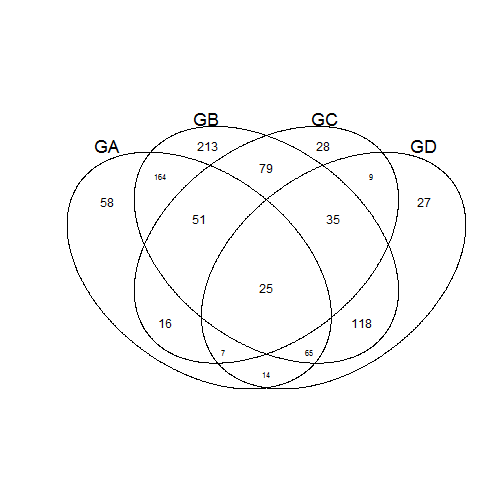
## Example using a list of item indexes belonging to the specified group.
GroupA2 <- which(geneNames %in% GroupA)
GroupB2 <- which(geneNames %in% GroupB)
GroupC2 <- which(geneNames %in% GroupC)
GroupD2 <- which(geneNames %in% GroupD)
input2 <- list(GA2 = GroupA2, GB2 = GroupB2, GC2 = GroupC2, GD2 = GroupD2)
venn(input2)
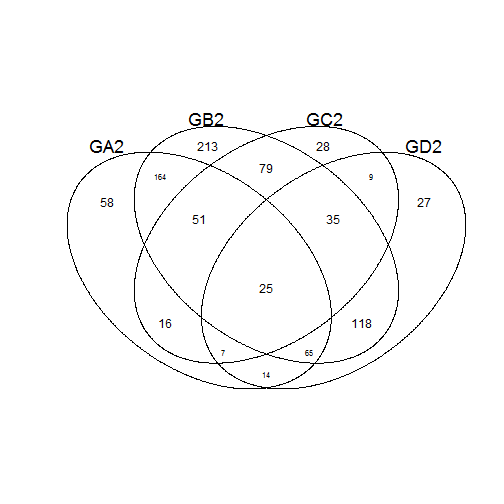
## Example using a data frame of indicator ('f'lag) columns
GroupA.f <- geneNames %in% GroupA
GroupB.f <- geneNames %in% GroupB
GroupC.f <- geneNames %in% GroupC
GroupD.f <- geneNames %in% GroupD
input.df <- data.frame(A = GroupA.f, B = GroupB.f, C = GroupC.f, D = GroupD.f)
head(input.df)
A B C D
1 TRUE TRUE TRUE FALSE
2 TRUE TRUE FALSE TRUE
3 FALSE TRUE FALSE FALSE
4 FALSE FALSE FALSE FALSE
5 FALSE TRUE FALSE FALSE
6 FALSE TRUE TRUE FALSE
venn(input.df)
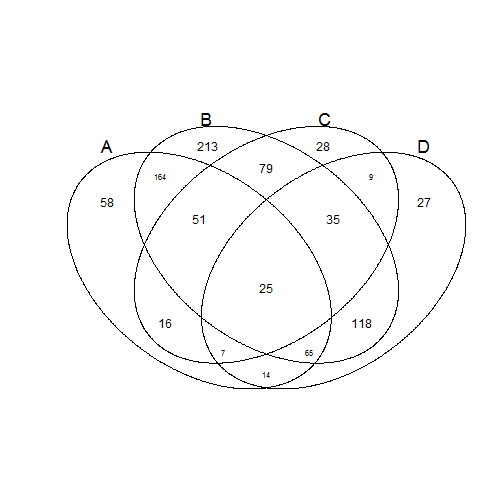
## Example to determine which elements are in A and B but not in C and D:
## first determine the universe, then form indicator columns and perform
## intersections on these. R allows using the set operations directly, but
## some might find this approach more intuitive.
universe <- unique(c(GroupA, GroupB, GroupC, GroupD))
GroupA.l <- universe %in% GroupA
GroupB.l <- universe %in% GroupB
GroupC.l <- universe %in% GroupC
GroupD.l <- universe %in% GroupD
## Genes that are in GroupA and in GroupB but not in GroupD (unification of
## sets III0 and II00 in the venn diagram:
universe[GroupA.l & GroupB.l & !GroupD.l]
[1] "LJNGD" "UQVQH" "MMHSM" "DLJVX" "KUOPR" "QOOLP" "OVBIQ" "FFDFP"
[9] "ISMFE" "VTFGZ" "OCSSQ" "NUSGS" "TEZQK" "CBDPU" "HAKQH" "OOIZX"
[17] "YUQMS" "BQWZM" "VNXBM" "BXSWW" "YTDTJ" "DAQDB" "EKSTZ" "PQSCL"
[25] "MTZSP" "SFAGP" "BOTQZ" "LBWHV" "ASWAY" "IVFVS" "UJCZN" "RWOHB"
[33] "VVLWB" "AWNWU" "AIGKN" "PQCLN" "IGIFJ" "ZSWHS" "LAFWD" "ALPXH"
[41] "ZOACU" "FZJAB" "ZKUEG" "PBXGH" "EAKES" "HSHVF" "ILWER" "NKDYR"
[49] "NPICS" "ANFOE" "SETBU" "TQYRT" "QUXJT" "UUEAJ" "DXUKG" "ZGQLP"
[57] "FCZWQ" "OXYHX" "WIYEM" "RYYYT" "WVCGX" "NOLXE" "NOQBP" "KLWMC"
[65] "LSHHB" "QIHLG" "FVTFO" "JZNPN" "KFYAR" "HCYRD" "XRXLW" "ROALB"
[73] "FTHWY" "HKGVI" "YNTXC" "RHFCM" "RAXOG" "ZQOLJ" "PSJSV" "ARFWR"
[81] "JTGPX" "GQCLK" "RPUDD" "NAPEH" "DADCB" "JRTWN" "HCZKQ" "SNGZB"
[89] "TZUVO" "NXMZV" "DTFUR" "VAVAE" "QQMLZ" "JGNQQ" "FHCUD" "ZGVYH"
[97] "JXJFA" "FKQJQ" "LLREW" "IPCYI" "WBJYA" "JNLHV" "PRLIX" "SXRJP"
[105] "DMUVQ" "CVAZE" "YZLDT" "TTEVZ" "XGFXY" "IDVML" "LHXLT" "ADWRG"
[113] "RKMBY" "YBHBB" "LJJIT" "VLFCS" "BBBZQ" "KZJVR" "RNBOI" "NUVPE"
[121] "RFJVT" "VPDIH" "KSRZY" "XVUJP" "IGBTO" "BNLXS" "BPKAV" "LPERU"
[129] "DXHLN" "YMMJE" "IIUWM" "GLDOR" "SRSZN" "ZFXIY" "AXOJE" "HEJOW"
[137] "OVNMM" "KGHKC" "YARXA" "TJQHL" "TTGCQ" "QXJGM" "TQBBD" "IXCRE"
[145] "HLHFX" "IWHBS" "RRHBX" "AZXXQ" "KOSXX" "HCJKY" "YIEWX" "VGSJD"
[153] "MYGOI" "YZTCW" "WXIZF" "GMJRT" "GHFDP" "LINFA" "RANSM" "SDWXZ"
[161] "BWXMW" "WKSIK" "OIGKP" "RTDBJ" "RTXSQ" "ZBLTC" "XSJUA" "KEOID"
[169] "FZLBM" "EBYBZ" "KKZPT" "ECLUE" "FNEMV" "WHZGV" "DYKBX" "HWXKY"
[177] "AWCHJ" "RZTGO" "NVHUR" "EZJKS" "USKOB" "UQRTP" "ETLNG" "CSQLL"
[185] "BAWFQ" "XZWJR" "BYCKD" "DGGUJ" "ESWWX" "BGXYC" "NDRLP" "HWYGQ"
[193] "MQCUS" "IOFIY" "XZCJZ" "STFLH" "DCKQJ" "VSTJX" "ZKMAU" "LJNHB"
[201] "QVXGB" "BVKBT" "OUBPF" "ZQIRR" "NIJXI" "SLYHJ" "UCDXR" "UHFJL"
[209] "ZUHMY" "POVCR" "MLTSH" "HRCGY" "ZVRDU" "SJWXH" "LJASB"
## Alternatively: construct a function to test for the pattern you want.
test <- function(x) (x %in% GroupA) & (x %in% GroupB) & !(x %in% GroupC)
universe[test(universe)]
[1] "BDCSG" "UQVQH" "MMHSM" "THMNP" "JNHNT" "DLJVX" "KUOPR" "QOOLP"
[9] "FFDFP" "ISMFE" "CBDPU" "HAKQH" "OOIZX" "YUQMS" "BQWZM" "VNXBM"
[17] "NOBXM" "WYAFV" "YTDTJ" "DAQDB" "EKSTZ" "PQSCL" "ZRVVD" "WDKCI"
[25] "CBJTV" "SFAGP" "BOTQZ" "LBWHV" "ASWAY" "IYAVE" "IVFVS" "UJCZN"
[33] "RWOHB" "VVLWB" "PQCLN" "IGIFJ" "ZSWHS" "AXVHG" "LAFWD" "ALPXH"
[41] "ZOACU" "FZJAB" "JDKWV" "ZKUEG" "PBXGH" "EAKES" "HSHVF" "ILWER"
[49] "NKDYR" "NPICS" "SPETC" "JMGTR" "SETBU" "OXSHI" "TQYRT" "KUGAT"
[57] "XFKNV" "UUEAJ" "JYQUN" "DXUKG" "KLODK" "ZGQLP" "FCZWQ" "OXYHX"
[65] "WIYEM" "RYYYT" "WVCGX" "NOLXE" "LSHHB" "QIHLG" "FVTFO" "KFYAR"
[73] "HCYRD" "XRXLW" "ROALB" "FTHWY" "YNTXC" "RHFCM" "RAXOG" "ZQOLJ"
[81] "PSJSV" "ARFWR" "JTGPX" "NTUDS" "RPUDD" "SCXPA" "NAPEH" "DADCB"
[89] "JRTWN" "SNGZB" "TZUVO" "NXMZV" "DTFUR" "KFULO" "VAVAE" "QQMLZ"
[97] "FKQJQ" "UYOAU" "IPCYI" "QRHVN" "PRLIX" "DMUVQ" "TTEVZ" "TADDM"
[105] "XGFXY" "IDVML" "LHXLT" "ADWRG" "RKMBY" "LJJIT" "KZJVR" "RNBOI"
[113] "NUVPE" "RFJVT" "AXDPZ" "XVUJP" "LYNLU" "FVMQZ" "IMHAR" "TTTXI"
[121] "DXHLN" "YMMJE" "FFKFJ" "IIUWM" "GLDOR" "SRSZN" "ZFXIY" "MJAPP"
[129] "AXOJE" "HEJOW" "OVNMM" "KGHKC" "YARXA" "TJQHL" "TTGCQ" "AVKPZ"
[137] "QXJGM" "TQBBD" "IXCRE" "HLHFX" "IWHBS" "RRHBX" "AZXXQ" "KOSXX"
[145] "HCJKY" "YIEWX" "VGSJD" "MYGOI" "YZTCW" "OSDDV" "WXIZF" "GMJRT"
[153] "GHFDP" "LINFA" "RANSM" "SDWXZ" "BWXMW" "OIGKP" "HIJYT" "RTDBJ"
[161] "QLBWK" "MWTGP" "XSJUA" "KEOID" "FZLBM" "THTUK" "HCNEP" "WXESQ"
[169] "KKZPT" "GJWXU" "ECLUE" "FNEMV" "WHZGV" "DYKBX" "MYEEF" "JVDHM"
[177] "HWXKY" "AQTJY" "RZTGO" "ZNYWF" "WLEYH" "EZJKS" "XFSVL" "KUDNT"
[185] "ETLNG" "CSQLL" "BAWFQ" "XZWJR" "BYCKD" "DGGUJ" "LEUNY" "AODBU"
[193] "ESWWX" "RMVJQ" "BGXYC" "CAKIG" "NDRLP" "MQCUS" "YDXFQ" "BMBJE"
[201] "IOFIY" "MROCC" "DCKQJ" "IBYUS" "VSTJX" "ZKMAU" "LJNHB" "SPAJS"
[209] "VMWPA" "QVXGB" "BVKBT" "OUBPF" "ZQIRR" "RYQPM" "NIJXI" "WSCZW"
[217] "NYYLH" "RJKNV" "RUGDL" "SLYHJ" "ITQRZ" "UCDXR" "BWYLK" "UHFJL"
[225] "HRCGY" "ZVRDU" "SJWXH" "JMNMK" "LJASB"
Further reading
gplots